Back in the year 2000 the Human Genome Project announced its completion. Yet the job wasn’t quite finished. Finally the last 8% has been read:
The human genome is, at long last, complete
Some of the properties, in bulk:
Total length:
Human Genome, in raw base-pairs: 3,054,815,472
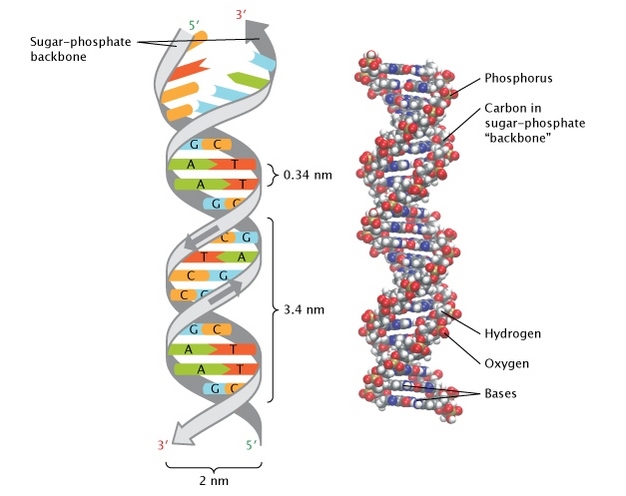
The Human Genetic Code is composed of 3 base-pair units, each base-pair having 4 possible choices, thus encoding a maximum of 4 X 4 X 4 = 64 possible choices per codon, therefore making it a six binary bit code. Six bit codes haven’t been used in computers for many years, but clearly it’s enough for biological systems. Bioscientists have successfully expanded the number of bases in the code in artificial systems, but it’s yet to be observed in Nature.
Total Human Codons: 1,018,271,824
Bits of Information, uncompressed: 6,109,630,944
Bytes (8-bit units): 763,703,868
Coded (bytes, produces mRNA): 351,751,368 (46.06%)
Repeats (bytes, repetitive “junk”): 411,952,500 (53.94%)
Curiously the actual code is not its theoretically maximum of a 4 X 4 X 4 = 64 possible choices. Instead the Genetic Code is highly redundant, with 20 amino acids and 1 stop code. Thus DNA codes roughly ~LN(21)/LN(2) = 4.4 bits per codon. Thanks to all the repetition and the code’s redundancy, the Human Genome can be significantly compressed. Roughly 260 megabytes of information. Once upon a time, that seemed like a lot of information – it is about 26,000 pages of the physical Encyclopedia Britannica – but it’s small compared to the data we store on our mobile devices these days.
An interesting addendum is that the number of genes identified increased slightly, but those ~20,000 genes encode about 70,000 proteins. More than one protein can be encoded by sequences, so the total information stored in DNA can be hard to pin down.

Good summary! I don’t know if I’d be so quick to dismiss that 53% of so-called repeats and junk though. Back when the genome project started that’s exactly what we thought of those sequences, but in the last decade or so that’s script has been flipped pretty thoroughly.
Like microbiomes, we don’t know a lot yet of those repetitive sequences, but what we are finding is that they are far from junk. They both seem to play vital yet still not well understood roles in regulation of genes’ expression. Likely more than just that too!
Ancient viruses, ancestral mutations, and who knows what else might be hidden there. You know the saying, one sophont’s genetic garbage is another’s treasure, or something like that ??
Hi Josh
A fair point. What it does mean is the repetitive segments are lower in information content and easily compressible. I don’t doubt they’re important – or at least non-deletrious to the genome – but they’re not “meaningful” in a ribosomal context and thus serve some other purpose.
Thank you for sharing your thoughts. I truly appreciate your efforts and I am waiting for your next write ups thanks
once again.